Feb 18th, 2016 Ontology Working Group Meeting: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
|||
| (83 intermediate revisions by the same user not shown) | |||
| Line 4: | Line 4: | ||
* Connection details: Join from PC, Mac, Linux, iOS or Android: https://zoom.us/j/627445291 | * Connection details: Join from PC, Mac, Linux, iOS or Android: https://zoom.us/j/627445291 | ||
[[File:Ontology WG Meeting 2-18-2016 video.mp4|framed|left|Ontology WG Meeting 2-18-2016 video]] | * Links to recordings: | ||
** [[File:Ontology WG Meeting 2-18-2016 video.mp4|framed|left|Ontology WG Meeting 2-18-2016 video]] | |||
** [[File:Ontology WG Meeting 2-18-2016 audio only.m4a|thumbnail|Ontology WG Meeting 2-18-2016 audio only]] | |||
* Attendees: AM, LC, CM, PJ, MAL, EA, GK, JE | |||
Agenda: | =Agenda: = | ||
==Amigo 2.0 issues== | |||
1. '''Display of relations''' | |||
''Currently, only the is_a, part_of and occurs_in relations are being displayed'' | |||
* See Tracker: [https://github.com/Planteome/planteome-ontology-browsers/issues/20 Relations are not being recognized and displayed in AmiGO 2.0 #20] | |||
* CM: its a priority for GO as well and they are working on it, although they have been short-handed, but are making it a priority for Planteome this month. | |||
** Could browse on OLS-beta: http://www.ebi.ac.uk/ols/beta/ontologies/to (''Note that this is only the live version to.owl- cannot view the crop specific ontologies there'') | |||
* Timeline: Would like to have this working correctly for the upcoming Release- ''by middle of March?'' | |||
* | *''Can we point the OLS and others like Bioportal to 'to-basic.obo'? Looks weird with the BFO and PLANTCHEBIROLE terms.'' | ||
2.''' Propagation of annotations:''' | |||
* | * The annotations should be moving correctly through the relations in the graph, even if they are not displayed | ||
** | ** CM: This is why the relations are being filtered. The old AmiGO would propagate everything, but now it can be set in the Planteome AmiGO 2.0 configuration. Generally, you only want the gene expression to propagate up through is_a and part_of | ||
''Need to look at the other PO relations such as has_part, participates_in, has_participant and for the TO, inheres_in and occurs_in and any others'' | |||
3. '''Display of CO terms with dash or colon.''' ''did not discuss this week'' | |||
* | * Example: cassava scale terms: http://dev.planteome.org/amigo/search/ontology | ||
==Revisions to the Equivalence Axioms for the TO== | |||
* Status of the BioCuration 2016 presentation and possible ICBO paper: Comparison of ontology mapping techniques to map traits (MAL) | |||
** bottleneck: needs of a collaborative revision of the design patterns (i.e. Equivalence Axioms) | |||
** | ===Summary of the patterns in use in TO currently:=== | ||
* ibp-crossproducts repo: [https://github.com/Planteome/ibp-crossproducts/wiki/Formal-definitions-in-TO Formal-definitions-in-TO] | |||
* see also: TO design pattern Google doc [[https://docs.google.com/document/d/1j5mUnkBrWIQJi9WX38POyuLrM1xDBD3GNGIyjv-zPRI/edit?usp=sharing]] | |||
** Contains some case information on formal definitions, and uses in trait definitions | |||
* Also: [https://docs.google.com/document/d/13DzXmSWjXtjv9Nv1iVtviTTGrGeIchomqiC4nIbbGu4/edit# Plant Trait Ontology Instances of 'occurs_in'] | |||
** CM: For documentation- there is also a YAML syntax - computable to autogenerate documentation- ''See the tutorial from Nov workshop'' | |||
* | ===Need to clarify when to use ''occurs_in'' and when to use ''inheres_in'' in the equivalence axioms=== | ||
** | * Currently 'occurs_in' is used in several of the classes such as content and composition, but according to BFO, it is technically supposed to be used to relate a process (continuant) to a material entity or immaterial entity (independent continuant). | ||
** currently occurs_in is used incorrectly towards non-processes?? | |||
- 'Content' classes currently have these three patterns: | |||
* Status of second year funding?? | * ‘amount’ (PATO:0000070) and inheres_in [CHEBI] (‘composition’(PATO:0000025)) | ||
* ‘amount’ (PATO:0000070) and inheres_in [CHEBI] and occurs_in [PO] | |||
* ‘composition’(PATO:0000025) and inheres_in [CHEBI] | |||
* Comments from CM: | |||
** ''We need to have the first two since sometimes it is unspecified- i.e.- it just in the plant itself- problem is that there is no entity in the ontologies to represent this.'' | |||
** ''The BFO definition (see below) of "occurs_in" does not fit the current understanding of PATO.'' | |||
** ''In a trait ontology you are interested in looking at the changes in a trait over time, but you are looking at a limit case at an instance in time.'' | |||
** It is OK to use 'occurs_in' as the issue is currently under discussion between PATO and BFO | |||
* BFO_0000066: occurs_in is specific to biological processes | |||
* See OntoBee: http://www.ontobee.org/ontology/RO?iri=http://purl.obolibrary.org/obo/BFO_0000066 | |||
* Definition: | |||
** b occurs_in c | |||
** b is a process | |||
** c is a material entity or immaterial entity | |||
** there exists a spatiotemporal region r | |||
** b occupies_spatiotemporal_region r | |||
** for all (t) if b exists_at t, then c exists_at t | |||
** there exist spatial regions s and s’, where & b spatially_projects_onto s at t | |||
** c is occupies_spatial_region s’ at t & s is a proper_continuant_part_of s’ at t | |||
* Paraphrase of definition: a relation between a process and an independent continuant, in which the process takes place entirely within the independent continuant | |||
* alternative term: unfolds in; unfolds_in; occurs_in | |||
===How to model count information: ''has_number_of'' vs ''amount''=== | |||
PATO vs. BFO | |||
* BFO wants to use ''has_number_of'' to describe count information, and PATO wants to use ''amount'' | |||
* BFO- annotate to the larger entity (having a number of) | |||
* PATO - smaller entity has an amount (or towards) | |||
* 'has_number_of' is a relational quality | |||
* CM: Some trait ontologies are using the older, more simple pattern PATO ''number (ie amount)'' that inheres in the thing that is being counted. | |||
** ''inheres_in'' the thing you're counting, ''occurs_in'' whatever larger entity | |||
* Example: To describe the number of seeds in a pod: quality = amount, entity = seed, occurs_in pod | |||
** don't use relational qualities (eg: has_number_of) | |||
** ''occurs_in'' == ''towards'' | |||
* amount can inhere in the object being counted, where as has_number_of, is a quality of the larger entity | |||
* how to describe "number_of" | |||
* has_number_of molecules, that's why has-number-of | |||
* We should annotate to the upper-level "molecular entity"; being consistent is more important than being correct (ontologically) | |||
e.g. iron molecular entity | |||
* concentration: has increased concentration in a mixture, either increased amount in a mixture | |||
=== Discussion of PATO ''Composition''=== | |||
* We are using composition (PATO_0000025) to describe the content classes, since amount actually means "number_of" | |||
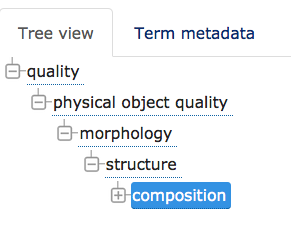
* Currently, PATO term composition (PATO_0000025) is subclass of structure (PATO_0000141), in the morphology branch of physical object quality. | |||
* composition (PATO_0000025): A single physical entity inhering in an bearer by virtue of the bearer's quantities or relative ratios of subparts. [ PATOC:GVG ] | |||
* See diagram: [[File:PATO composition.png|thumbnail|PATO_composition.png]] | |||
* composition vs. amount: composition (PATO) cannot be used for non-morphology things (like blood, or sap) | |||
** composition is in the structure branch, and should be renamed "structural composition" or "structure composition" | |||
** - ''Need to open tracker for this'' | |||
* PJ: suggestion to move composition out of subclass of structure, need to be able to describe the composition of entities such as ''xylem sap''. | |||
** this would make it a physical object quality | |||
* Comment from BS: The content /composition terms should be a subclass of the morphology of the structure, rather than being in a separate branch. ''This would actually make sense in a way, since it is in the physical object quality branch. | |||
** For example: | |||
* View on PATO on http://www.ebi.ac.uk/ols/ontologies/pato | |||
== Display of the Crop Ontologies in AmiGO2 == | |||
* Crop-specific names have been added to the rice, cassava and lentil files | |||
* Names should be in lower case | |||
==Visit to IRRI== | |||
* Monday March 7th to Friday March 11th | |||
* Goals: TD revision and data annotation- MAL, and Leo will be working on trait dictionary | |||
* Austin- send me the contact info | |||
* all 3 - should be contacting each other as a group | |||
==Status of second year funding== | |||
* Bioversity is getting impatient, PJ will follow up with NSF and find out | |||
= Upcoming Meetings and Workshops= | |||
==[http://www.phenotypercn.org/?page_id=2750 Phenotype RCN meeting February 26-28, 2016] == | |||
* PJ is going | |||
==[https://www.isb-sib.ch/events/biocuration2016/home Biocuration 2016, April 10th-14th,2016; Geneva, Switzerland]== | |||
* MAL is going | |||
** [https://www.isb-sib.ch/events/biocuration2016/key-dates key dates] | |||
** Abstract submitted: https://easychair.org/conferences/submission.cgi?a=10690939;submission=2641703 | |||
==[http://blog.garnetcommunity.org.uk/garnetegenis-workshop-integrating-large-data-plant-science-big-data-discovery/ GARNet/Egenis Workshop: Integrating Large Data into Plant Science], April 21st-22nd 2016, Dartington Hall, Totnes, Devon== | |||
* Elizabeth and George are going, EA will present Planteome as part of her talk | |||
== Meeting in Montpellier, May 2016- website??== | |||
==[http://www.bio-ontologies.org.uk/call-for-participation BioOntologies SIG] of the [http://www.iscb.org/ismb2016 Intelligent Systems for Molecular Biology (ISMB)]; July 8-12, 2016, Orlando, Florida == | |||
* Dates: July 8th and 9th, with July 9th being the “Phenotype Day”, focused on the systematic description of phenotypes. | |||
** Short papers, up to 4 pages (will be published in JBMS) | |||
** Poster abstracts, up to 1 page | |||
** Flash updates, up to 1 page | |||
== [http://icbo.cgrb.oregonstate.edu/ 7th International Conference on Biological Ontology and BioCreative 2016] Aug 1st to 4th, Corvallis, OR== | |||
* Link to program: [http://icbo.cgrb.oregonstate.edu/node/20 ICBO + BioCreative Program] | |||
* Link to Easy Chair site: https://easychair.org/conferences/conference_info.cgi?a=10776589 | |||
Latest revision as of 22:50, 7 July 2016
Planteome Ontology WG Zoom Meeting
- Date: Thursday Feb 18th, 2016
- Time: 8:15am PST (GMT-8)
- Connection details: Join from PC, Mac, Linux, iOS or Android: https://zoom.us/j/627445291
- Links to recordings:
- Attendees: AM, LC, CM, PJ, MAL, EA, GK, JE
Agenda:
Amigo 2.0 issues
1. Display of relations Currently, only the is_a, part_of and occurs_in relations are being displayed
- See Tracker: Relations are not being recognized and displayed in AmiGO 2.0 #20
- CM: its a priority for GO as well and they are working on it, although they have been short-handed, but are making it a priority for Planteome this month.
- Could browse on OLS-beta: http://www.ebi.ac.uk/ols/beta/ontologies/to (Note that this is only the live version to.owl- cannot view the crop specific ontologies there)
- Timeline: Would like to have this working correctly for the upcoming Release- by middle of March?
- Can we point the OLS and others like Bioportal to 'to-basic.obo'? Looks weird with the BFO and PLANTCHEBIROLE terms.
2. Propagation of annotations:
- The annotations should be moving correctly through the relations in the graph, even if they are not displayed
- CM: This is why the relations are being filtered. The old AmiGO would propagate everything, but now it can be set in the Planteome AmiGO 2.0 configuration. Generally, you only want the gene expression to propagate up through is_a and part_of
Need to look at the other PO relations such as has_part, participates_in, has_participant and for the TO, inheres_in and occurs_in and any others
3. Display of CO terms with dash or colon. did not discuss this week
- Example: cassava scale terms: http://dev.planteome.org/amigo/search/ontology
Revisions to the Equivalence Axioms for the TO
- Status of the BioCuration 2016 presentation and possible ICBO paper: Comparison of ontology mapping techniques to map traits (MAL)
- bottleneck: needs of a collaborative revision of the design patterns (i.e. Equivalence Axioms)
Summary of the patterns in use in TO currently:
- ibp-crossproducts repo: Formal-definitions-in-TO
- see also: TO design pattern Google doc [[1]]
- Contains some case information on formal definitions, and uses in trait definitions
- Also: Plant Trait Ontology Instances of 'occurs_in'
- CM: For documentation- there is also a YAML syntax - computable to autogenerate documentation- See the tutorial from Nov workshop
Need to clarify when to use occurs_in and when to use inheres_in in the equivalence axioms
- Currently 'occurs_in' is used in several of the classes such as content and composition, but according to BFO, it is technically supposed to be used to relate a process (continuant) to a material entity or immaterial entity (independent continuant).
- currently occurs_in is used incorrectly towards non-processes??
- 'Content' classes currently have these three patterns:
- ‘amount’ (PATO:0000070) and inheres_in [CHEBI] (‘composition’(PATO:0000025))
- ‘amount’ (PATO:0000070) and inheres_in [CHEBI] and occurs_in [PO]
- ‘composition’(PATO:0000025) and inheres_in [CHEBI]
- Comments from CM:
- We need to have the first two since sometimes it is unspecified- i.e.- it just in the plant itself- problem is that there is no entity in the ontologies to represent this.
- The BFO definition (see below) of "occurs_in" does not fit the current understanding of PATO.
- In a trait ontology you are interested in looking at the changes in a trait over time, but you are looking at a limit case at an instance in time.
- It is OK to use 'occurs_in' as the issue is currently under discussion between PATO and BFO
- BFO_0000066: occurs_in is specific to biological processes
- See OntoBee: http://www.ontobee.org/ontology/RO?iri=http://purl.obolibrary.org/obo/BFO_0000066
- Definition:
- b occurs_in c
- b is a process
- c is a material entity or immaterial entity
- there exists a spatiotemporal region r
- b occupies_spatiotemporal_region r
- for all (t) if b exists_at t, then c exists_at t
- there exist spatial regions s and s’, where & b spatially_projects_onto s at t
- c is occupies_spatial_region s’ at t & s is a proper_continuant_part_of s’ at t
- Paraphrase of definition: a relation between a process and an independent continuant, in which the process takes place entirely within the independent continuant
- alternative term: unfolds in; unfolds_in; occurs_in
How to model count information: has_number_of vs amount
PATO vs. BFO
- BFO wants to use has_number_of to describe count information, and PATO wants to use amount
- BFO- annotate to the larger entity (having a number of)
- PATO - smaller entity has an amount (or towards)
- 'has_number_of' is a relational quality
- CM: Some trait ontologies are using the older, more simple pattern PATO number (ie amount) that inheres in the thing that is being counted.
- inheres_in the thing you're counting, occurs_in whatever larger entity
- Example: To describe the number of seeds in a pod: quality = amount, entity = seed, occurs_in pod
- don't use relational qualities (eg: has_number_of)
- occurs_in == towards
- amount can inhere in the object being counted, where as has_number_of, is a quality of the larger entity
- how to describe "number_of"
- has_number_of molecules, that's why has-number-of
- We should annotate to the upper-level "molecular entity"; being consistent is more important than being correct (ontologically)
e.g. iron molecular entity
- concentration: has increased concentration in a mixture, either increased amount in a mixture
Discussion of PATO Composition
- We are using composition (PATO_0000025) to describe the content classes, since amount actually means "number_of"
- Currently, PATO term composition (PATO_0000025) is subclass of structure (PATO_0000141), in the morphology branch of physical object quality.
- composition (PATO_0000025): A single physical entity inhering in an bearer by virtue of the bearer's quantities or relative ratios of subparts. [ PATOC:GVG ]
- composition vs. amount: composition (PATO) cannot be used for non-morphology things (like blood, or sap)
- composition is in the structure branch, and should be renamed "structural composition" or "structure composition"
- - Need to open tracker for this
- PJ: suggestion to move composition out of subclass of structure, need to be able to describe the composition of entities such as xylem sap.
- this would make it a physical object quality
- Comment from BS: The content /composition terms should be a subclass of the morphology of the structure, rather than being in a separate branch. This would actually make sense in a way, since it is in the physical object quality branch.
- For example:
- View on PATO on http://www.ebi.ac.uk/ols/ontologies/pato
Display of the Crop Ontologies in AmiGO2
- Crop-specific names have been added to the rice, cassava and lentil files
- Names should be in lower case
Visit to IRRI
- Monday March 7th to Friday March 11th
- Goals: TD revision and data annotation- MAL, and Leo will be working on trait dictionary
- Austin- send me the contact info
- all 3 - should be contacting each other as a group
Status of second year funding
- Bioversity is getting impatient, PJ will follow up with NSF and find out
Upcoming Meetings and Workshops
Phenotype RCN meeting February 26-28, 2016
- PJ is going
Biocuration 2016, April 10th-14th,2016; Geneva, Switzerland
- MAL is going
GARNet/Egenis Workshop: Integrating Large Data into Plant Science, April 21st-22nd 2016, Dartington Hall, Totnes, Devon
- Elizabeth and George are going, EA will present Planteome as part of her talk
Meeting in Montpellier, May 2016- website??
BioOntologies SIG of the Intelligent Systems for Molecular Biology (ISMB); July 8-12, 2016, Orlando, Florida
- Dates: July 8th and 9th, with July 9th being the “Phenotype Day”, focused on the systematic description of phenotypes.
- Short papers, up to 4 pages (will be published in JBMS)
- Poster abstracts, up to 1 page
- Flash updates, up to 1 page
7th International Conference on Biological Ontology and BioCreative 2016 Aug 1st to 4th, Corvallis, OR
- Link to program: ICBO + BioCreative Program
- Link to Easy Chair site: https://easychair.org/conferences/conference_info.cgi?a=10776589