Difference between revisions of "Thursday March 5th, 2015"
(→C. Update from IT group- Data Store, AmiGO2, Justin Elser:) |
(→A. General Comments and Updates:) |
||
| (43 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
Who: EA, LC, JE, JP Not available: PJ, BS, GG, EZ, ST | Who: EA, LC, JE, JP Not available: PJ, BS, GG, EZ, ST | ||
| − | + | * Link to recording- Video: [[File:AllHands 3-5-15.mp4|thumbnail|AllHands 3-5-15.mp4]] | |
| + | * Link to recording- Audio: [[File:AllHands Audio 3-5-15.m4a|thumbnail|AllHands Audio 3-5-15.m4a]] | ||
== A. General Comments and Updates:== | == A. General Comments and Updates:== | ||
| Line 12: | Line 13: | ||
* PJ will circulate final version of the proposal in a cleaned-up format | * PJ will circulate final version of the proposal in a cleaned-up format | ||
| − | ''- EA- It would be very useful to have a copy of the 'official abstract' to share with partners or colleagues.'' | + | ''- EA- It would be very useful to have a copy of the 'official abstract' to share with partners or colleagues. See link to award on NSF site: http://www.nsf.gov/awardsearch/showAward?AWD_ID=1340112'' |
| − | ''- LC will send EA the NSF logos to use with the announcement on the Crop Ontology page'' | + | ''- LC will send EA the NSF logos to use with the announcement on the Crop Ontology page'' Done! |
* Progress on opening new Planteome-related positions in the Jaiswal Lab (software developer, curator)- | * Progress on opening new Planteome-related positions in the Jaiswal Lab (software developer, curator)- | ||
| Line 24: | Line 25: | ||
* A test version of the AmiGO 2.2.3 browser is up and running at [http://dev.planteome.org/amigo Amigo 2.0 on Planteome.dev]. | * A test version of the AmiGO 2.2.3 browser is up and running at [http://dev.planteome.org/amigo Amigo 2.0 on Planteome.dev]. | ||
- It currently just has a standard set of ontologies and a small subset of annotations. | - It currently just has a standard set of ontologies and a small subset of annotations. | ||
| − | |||
| − | |||
| − | + | - To navigate: go to http://dev.planteome.org/amigo/search/ontology | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| + | * Note: I was unable to get it to load with the ncbi taxon ontology like GO has because the loading program ran out of memory. | ||
| + | ''Possibly amy be better to just load a branch of the NCBI taxonomy, for the plants- we do not really need the whole thing.'' | ||
| − | + | * Next steps are to work on the theming and to clean up the list of the ontologies: | |
| − | * | + | |
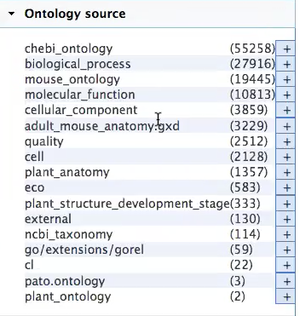
| + | ''Currently, this is the list of ontologies that are loaded:'' | ||
| + | |||
| + | [[File:AmiGO2 Planteome dev.png|thumbnail|AmiGO2 Planteome dev.png]] | ||
| + | |||
| + | Links: | ||
| + | * http://purl.obolibrary.org/obo/go/extensions/go-plus.owl | ||
| + | * [http://purl.obolibrary.org/obo/ncbitaxon.owl%20\ NCBI Taxonomy ] | ||
| + | * http://purl.obolibrary.org/obo/cl/cl-basic.owl \ | ||
| + | * [http://purl.obolibrary.org/obo/eco.owl%20\ Evidence Code Ontology] | ||
| + | * http://purl.obolibrary.org/obo/go/extensions/gorel.owl \ | ||
| + | * http://purl.obolibrary.org/obo/ma.owl \ | ||
| + | * http://purl.obolibrary.org/obo/pato.owl \ | ||
| + | * http://purl.obolibrary.org/obo/po.owl \ | ||
| + | * http://purl.obolibrary.org/obo/emap.owl \ | ||
| + | * http://purl.obolibrary.org/obo/chebi.owl \ | ||
| + | * http://purl.obolibrary.org/obo/uberon/basic.owl \ | ||
| + | * http://purl.obolibrary.org/obo/go/extensions/go-modules-annotations.owl | ||
| + | |||
| + | * Priority for us will be refining the list of the ontologies: Do not need the following: | ||
| + | ** Mouse_ontology | ||
| + | **adult_mouse_anatomy | ||
| + | * Need to include the Trait Ontology | ||
| + | |||
| + | *Also need to check on the versions of the ontologies being loaded from the purls above- may be able to modify to use our verisons. | ||
| + | |||
| + | * Load time for the ontologies is about 2 hours and the test set of annotations loaded in a few minutes. | ||
* JE is working with Chris Sullivan to figure out a way to be able to do the load from one of the compute nodes in the cluster as they have much more RAM. | * JE is working with Chris Sullivan to figure out a way to be able to do the load from one of the compute nodes in the cluster as they have much more RAM. | ||
| − | * | + | * For more information, please see the notes at: [[Data_storage_and_AmiGO2_Working_Group]] |
== D. Update from AISO/BisQue group- Justin Preece:== | == D. Update from AISO/BisQue group- Justin Preece:== | ||
| − | * | + | * Bisque (http://bioimage.ucsb.edu/bisque) is the Image Annotation (IA) of iPlant, and is the platform we will be using for the Planteome |
| + | * Last group meeting was Feb 12th, [[Image_Annotation_Working_Group_Meetings]] | ||
| + | * Setting up development environment for the two graduate students who work with Sinisa | ||
| + | * Starting to work on the IA modules with in Bisque | ||
| + | * Bisque has some aspects of segmentation, but it is limited, compared to the AISO desktop tool | ||
| + | * Initial focus is to build a basic user interface that allows single segmentation input- it sends it out to the Bisque server and returns a segmented image. Not interactive- done over the network | ||
| + | * Bisque lacks the software architecture to do interactive segmentation, over the network. | ||
| + | * Bisque developers are working on a more synchronous interactive mode (independent of our project) | ||
| + | * [https://docs.google.com/document/d/14i3ED8cAX6-qwdpko8eV8aQrwplLRfLHZ93dZYpjZu8/edit Draft doc of the specs] is linked to the wiki page. | ||
| − | + | ===Continuing the discussion about the Generation of Sample Images from last month - DWS=== | |
| − | + | ||
** Need to generate the labeled images- initially using AISO | ** Need to generate the labeled images- initially using AISO | ||
* Priorities would be images of leaf types, flowers, root and stem sections | * Priorities would be images of leaf types, flowers, root and stem sections | ||
| − | |||
| − | |||
* Will go along with the glossary of anatomy terms based on the PO | * Will go along with the glossary of anatomy terms based on the PO | ||
| + | * Recent AVATOL grant meeting: http://avatol.org/; http://currents.plos.org/treeoflife/avatol/; http://currents.plos.org/treeoflife/article/next-generation-phenomics-for-the-tree-of-life/ | ||
| + | ** Generating leaf clearance images | ||
| + | * DWS has a large number of images from the plant anatomy textbook - will send us the link or the CD, working on the 2nd edition now | ||
| + | * DWS owns them so it is not a problem, part of the laboratory exercises on a CD for the book | ||
| + | * Will need to make sure it is ok with the publisher- Wiley | ||
| + | * Machine learning will happen in Phase 3- can develop the needed images | ||
== E. Update from Ontologies Working group:== | == E. Update from Ontologies Working group:== | ||
| − | + | * Updates from recent weekly meetings: [http://wiki.planteome.org/index.php?title=Ontology_Working_Group_Meetings-_Feb_2015#3._Panzea_Project:_Curating_Maize_Diversity_with_MaizeGDB Panzea project], [http://wiki.planteome.org/index.php?title=Ontology_Working_Group_Meetings-_Feb_2015#1._Review_of_the_Crop_Ontology_Structure Crop Ontology] | |
| − | + | * Barry presented the Planteome to the United Nations | |
| − | * Next meeting is Friday Mar 6th, 2015 8:30am PST (GMT-8) | + | * [https://rd-alliance.org/groups/wheat-data-interoperability-wg.html Wheat Data Interoperability Working Group] at the RDA Plenary in San Diego next week. Presenting the Recommended data standards (ie: cookbook) and results of the ontology survey. |
| + | * Next meeting is Friday Mar 6th, 2015 8:30am PST (GMT-8)- Rosemary Shrestha - Data manager at CIMMYT, curator of the Maize trait Ontology | ||
| + | * New team member Leo Valette is joining the IBP and CO in Montpellier. | ||
===Update on Position advertisements:=== | ===Update on Position advertisements:=== | ||
| Line 66: | Line 96: | ||
* Oregon State University Advertisement: posted on various sites: http://aspb.site-ym.com/networking/apply_now.aspx?view=2&id=276580 | * Oregon State University Advertisement: posted on various sites: http://aspb.site-ym.com/networking/apply_now.aspx?view=2&id=276580 | ||
| − | == | + | ==Other Comments== |
| − | + | DWS: Comments from colleagues at NYU, on the GO and how it does not really tell you too much in reality. Ideally the Planteome will link the genes and functions and processes, to the final outcome of the plant development and plant structures. | |
| − | + | * Suggestion is to do more links between the GO and PO- knowledge is there, but has not been exploited. Moving into plant development and the traits | |
| − | + | * Example shown by PJ- network graph, that shows the link genes, functions and processes, to the final outcome of the plant development and plant structures. | |
| − | + | * Sun gear type diagrams | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | * | + | |
| − | + | ||
| − | * | + | |
| − | * | + | |
Latest revision as of 16:42, 15 April 2015
Who: EA, LC, JE, JP Not available: PJ, BS, GG, EZ, ST
- Link to recording- Video: File:AllHands 3-5-15.mp4
- Link to recording- Audio: File:AllHands Audio 3-5-15.m4a
Contents
A. General Comments and Updates:
- Schedule for Monthly All-Hands meetings
- We will do a new doodle poll- LC
- Planteome Wiki page has been set up with password protection (if you are reading this, it must be working :-))
From last meeting...
- PJ will circulate final version of the proposal in a cleaned-up format
- EA- It would be very useful to have a copy of the 'official abstract' to share with partners or colleagues. See link to award on NSF site: http://www.nsf.gov/awardsearch/showAward?AWD_ID=1340112
- LC will send EA the NSF logos to use with the announcement on the Crop Ontology page Done!
- Progress on opening new Planteome-related positions in the Jaiswal Lab (software developer, curator)-
- position is open, being advertised on various sites: Planteome blog, ASPB Careers
C. Update from IT group- Data Store, AmiGO2, Justin Elser:
- A test version of the AmiGO 2.2.3 browser is up and running at Amigo 2.0 on Planteome.dev.
- It currently just has a standard set of ontologies and a small subset of annotations.
- To navigate: go to http://dev.planteome.org/amigo/search/ontology
- Note: I was unable to get it to load with the ncbi taxon ontology like GO has because the loading program ran out of memory.
Possibly amy be better to just load a branch of the NCBI taxonomy, for the plants- we do not really need the whole thing.
- Next steps are to work on the theming and to clean up the list of the ontologies:
Currently, this is the list of ontologies that are loaded:
Links:
- http://purl.obolibrary.org/obo/go/extensions/go-plus.owl
- NCBI Taxonomy
- http://purl.obolibrary.org/obo/cl/cl-basic.owl \
- Evidence Code Ontology
- http://purl.obolibrary.org/obo/go/extensions/gorel.owl \
- http://purl.obolibrary.org/obo/ma.owl \
- http://purl.obolibrary.org/obo/pato.owl \
- http://purl.obolibrary.org/obo/po.owl \
- http://purl.obolibrary.org/obo/emap.owl \
- http://purl.obolibrary.org/obo/chebi.owl \
- http://purl.obolibrary.org/obo/uberon/basic.owl \
- http://purl.obolibrary.org/obo/go/extensions/go-modules-annotations.owl
- Priority for us will be refining the list of the ontologies: Do not need the following:
- Mouse_ontology
- adult_mouse_anatomy
- Need to include the Trait Ontology
- Also need to check on the versions of the ontologies being loaded from the purls above- may be able to modify to use our verisons.
- Load time for the ontologies is about 2 hours and the test set of annotations loaded in a few minutes.
- JE is working with Chris Sullivan to figure out a way to be able to do the load from one of the compute nodes in the cluster as they have much more RAM.
- For more information, please see the notes at: Data_storage_and_AmiGO2_Working_Group
D. Update from AISO/BisQue group- Justin Preece:
- Bisque (http://bioimage.ucsb.edu/bisque) is the Image Annotation (IA) of iPlant, and is the platform we will be using for the Planteome
- Last group meeting was Feb 12th, Image_Annotation_Working_Group_Meetings
- Setting up development environment for the two graduate students who work with Sinisa
- Starting to work on the IA modules with in Bisque
- Bisque has some aspects of segmentation, but it is limited, compared to the AISO desktop tool
- Initial focus is to build a basic user interface that allows single segmentation input- it sends it out to the Bisque server and returns a segmented image. Not interactive- done over the network
- Bisque lacks the software architecture to do interactive segmentation, over the network.
- Bisque developers are working on a more synchronous interactive mode (independent of our project)
- Draft doc of the specs is linked to the wiki page.
Continuing the discussion about the Generation of Sample Images from last month - DWS
- Need to generate the labeled images- initially using AISO
- Priorities would be images of leaf types, flowers, root and stem sections
- Will go along with the glossary of anatomy terms based on the PO
- Recent AVATOL grant meeting: http://avatol.org/; http://currents.plos.org/treeoflife/avatol/; http://currents.plos.org/treeoflife/article/next-generation-phenomics-for-the-tree-of-life/
- Generating leaf clearance images
- DWS has a large number of images from the plant anatomy textbook - will send us the link or the CD, working on the 2nd edition now
- DWS owns them so it is not a problem, part of the laboratory exercises on a CD for the book
- Will need to make sure it is ok with the publisher- Wiley
- Machine learning will happen in Phase 3- can develop the needed images
E. Update from Ontologies Working group:
- Updates from recent weekly meetings: Panzea project, Crop Ontology
- Barry presented the Planteome to the United Nations
- Wheat Data Interoperability Working Group at the RDA Plenary in San Diego next week. Presenting the Recommended data standards (ie: cookbook) and results of the ontology survey.
- Next meeting is Friday Mar 6th, 2015 8:30am PST (GMT-8)- Rosemary Shrestha - Data manager at CIMMYT, curator of the Maize trait Ontology
- New team member Leo Valette is joining the IBP and CO in Montpellier.
Update on Position advertisements:
- Crop Ontology position is closed, interviews are March 19-20th
- Oregon State University Advertisement: posted on various sites: http://aspb.site-ym.com/networking/apply_now.aspx?view=2&id=276580
Other Comments
DWS: Comments from colleagues at NYU, on the GO and how it does not really tell you too much in reality. Ideally the Planteome will link the genes and functions and processes, to the final outcome of the plant development and plant structures.
- Suggestion is to do more links between the GO and PO- knowledge is there, but has not been exploited. Moving into plant development and the traits
- Example shown by PJ- network graph, that shows the link genes, functions and processes, to the final outcome of the plant development and plant structures.
- Sun gear type diagrams