Thursday March 5th, 2015
Who: EA, LC, JE, JP Not available: PJ, BS, GG, EZ, ST
Link to recording: File:AllHands 3-5-15.mp4
A. General Comments and Updates:
- Schedule for Monthly All-Hands meetings
- We will do a new doodle poll- LC
- Planteome Wiki page has been set up with password protection (if you are reading this, it must be working :-))
From last meeting...
- PJ will circulate final version of the proposal in a cleaned-up format
- EA- It would be very useful to have a copy of the 'official abstract' to share with partners or colleagues. See link to award on NSF site: http://www.nsf.gov/awardsearch/showAward?AWD_ID=1340112
- LC will send EA the NSF logos to use with the announcement on the Crop Ontology page
- Progress on opening new Planteome-related positions in the Jaiswal Lab (software developer, curator)-
- position is open, being advertised on various sites: Planteome blog, ASPB Careers
C. Update from IT group- Data Store, AmiGO2, Justin Elser:
- A test version of the AmiGO 2.2.3 browser is up and running at Amigo 2.0 on Planteome.dev.
- It currently just has a standard set of ontologies and a small subset of annotations.
- To navigate: go to http://dev.planteome.org/amigo/search/ontology
- Note: I was unable to get it to load with the ncbi taxon ontology like GO has because the loading program ran out of memory.
Possibly amy be better to just load a branch of the NCBI taxonomy, for the plants- we do not really need the whole thing.
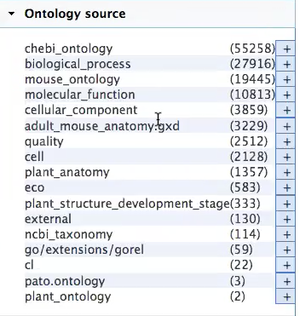
- Next steps are to work on the theming and to clean up the list of the ontologies:
Currently, this is the list of ontologies that are loaded:
Links:
- http://purl.obolibrary.org/obo/go/extensions/go-plus.owl
- NCBI Taxonomy
- http://purl.obolibrary.org/obo/cl/cl-basic.owl \
- Evidence Code Ontology
- http://purl.obolibrary.org/obo/go/extensions/gorel.owl \
- http://purl.obolibrary.org/obo/ma.owl \
- http://purl.obolibrary.org/obo/pato.owl \
- http://purl.obolibrary.org/obo/po.owl \
- http://purl.obolibrary.org/obo/emap.owl \
- http://purl.obolibrary.org/obo/chebi.owl \
- http://purl.obolibrary.org/obo/uberon/basic.owl \
- http://purl.obolibrary.org/obo/go/extensions/go-modules-annotations.owl
- Priority for us will be refining the list of the ontologies: Do not need the following:
- Mouse_ontology
- adult_mouse_anatomy
- Need to include the Trait Ontology
- Also need to check on the versions of the ontologies being loaded from the purls above- may be able to modify to use our verisons.
- Load time for the ontologies is about 2 hours and the test set of annotations loaded in a few minutes.
- JE is working with Chris Sullivan to figure out a way to be able to do the load from one of the compute nodes in the cluster as they have much more RAM.
- For more information, please see the notes at: Data_storage_and_AmiGO2_Working_Group
D. Update from AISO/BisQue group- Justin Preece:
- Bisque (http://bioimage.ucsb.edu/bisque) is the Image Annotation (IA) of iPlant, and is the platform we will be using for the Planteome
- Last group meeting was Feb 12th, Image_Annotation_Working_Group_Meetings
- Setting up development environment for the two graduate students who work with Sinisa
- Starting to work on the IA modules with in Bisque
- Bisque has some aspects of segmentation, but it is limited, compared to the AISO desktop tool
- Initial focus is to build a basic user interface that allows single segmentation input- it sends it out to the Bisque server and returns a segmented image. Not interactive- done over the network
- Bisque lacks the software architecture to do interactive segmentation, over the network.
- Bisque developers are working on a more synchronous interactive mode (independent of our project)
From last month's meeting:
- Generation of Sample Images - DWS
- Need to generate the labeled images- initially using AISO
- Priorities would be images of leaf types, flowers, root and stem sections
- DWS has a large number of images from the plant anatomy textbook - will send us the link
- DWS owns them so it is not a problem, part of the laboratory exercises on a CD for the book
- Will go along with the glossary of anatomy terms based on the PO
E. Update from Ontologies Working group:
Who is involved: EA, BS, DWS, LC, PJ, others TBA
- Next meeting is Friday Mar 6th, 2015 8:30am PST (GMT-8)
Update on Position advertisements:
- Crop Ontology position is closed, interviews are March 19-20th
- Oregon State University Advertisement: posted on various sites: http://aspb.site-ym.com/networking/apply_now.aspx?view=2&id=276580
F. Position Paper or "White Paper"
- In order to state our plans and cultivate our ideas, we should move quickly to produce a Position Paper or "White Paper".
- We do not want to lose out on ideas and concepts that we have proposed- others may take the credit and publish it first
- We should put forward our plans and stance, and what we expect to produce, rather than discussing it later in the project
- Possible places: Plant Methods, Nature Plants, Nature Methods, New Phytologist
- Will help us crystallize our ideas, goals and plans
Goals:
- Build a community consensus on widespread acceptance and availability, invite discussions
- Define the vocabularies and terms such as phenotypes, etc
- What level of phenotypes are we looking at: molecular aspects, function, processes, location, etc
- Any evaluated characteristic of an entity can considered a phenotype