Planteome Image Annotation Working Group: Difference between revisions
| Line 378: | Line 378: | ||
* Complete deployment and config of Planteome Integrated Dev server - JP, JE, RK | * Complete deployment and config of Planteome Integrated Dev server - JP, JE, RK | ||
* Define integration points between client and server software - RK, XX | * Define integration points between client and server software - RK, XX | ||
: | |||
=Conferences and Workshops= | =Conferences and Workshops= | ||
Please see: [[Planteome_Outreach]] | Please see: [[Planteome_Outreach]] | ||
Revision as of 18:21, 25 September 2015
Goals and Objectives
Detailed Software Specification
Tentative Development Timeline (based on current staffing and resources)
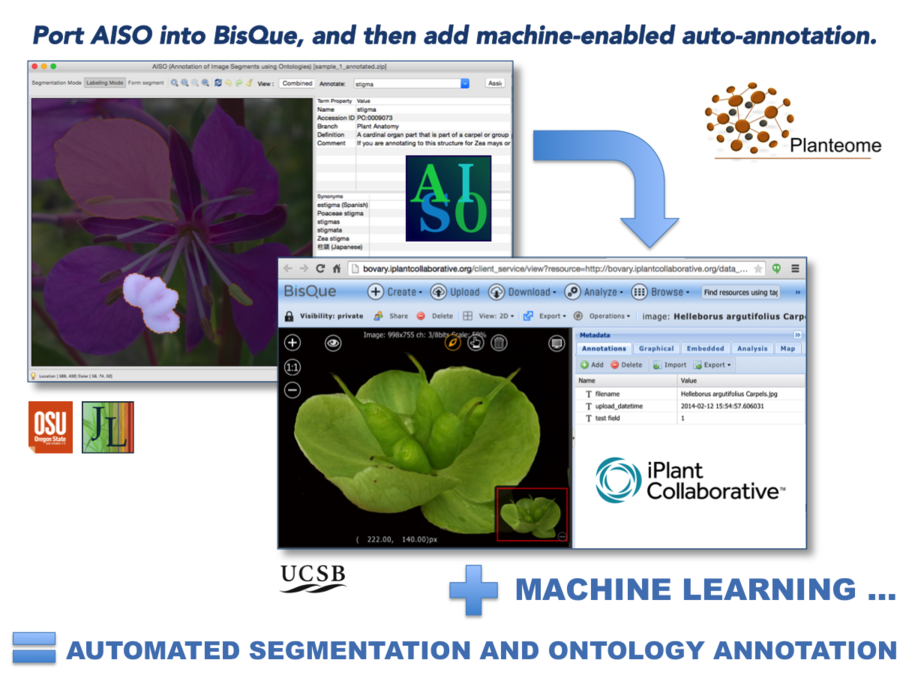
More about AISO, plus downloads.
Phase 1: Spring-Summer 2015
Objective: Implement asynchronous user-directed graph cuts in the Bisque environment.
Summary
- Build a Bisque module that allows the user to “mark up” an uploaded image, submit that markup to the Bisque server for graph cut segmentation, and receive the segmented results as a layered annotation.
- Build and integrate a separate feature into the metadata space to allow annotation of image segments with ontology terms (eventually provided remotely via web service.)
Phase 2: Summer-Fall-Winter 2015
Objective: Implement “wired” ontology metadata. Interactive, single-session graph cut segmentation of multiple, potentially overlapping segments on one image.
Summary:
- Allow the user to provide multiple mark-up lines and perform one or more segmentations on an image in near-real time, during a single user session. Will also allow revision tracking for multiple undo-redo steps as required by the user.
- Enhance the segmentation feature to render and manage multiple segment objects on a single image, with appropriate overlap, translucency, and inter-segment navigation capabilities.
Phase 3: 2016-2017
Objective(s):
- Implement machine-learning (“active learning”) algorithms to optimize segmentation and to auto-annotate plant images
- Optimize for very large, hi-res images (tiling? scaling?)
- Enable use of localized ontology reference data (a la custom ontologies provided by the user, or in an uploaded file)
Participants
- Jaiswal Lab (OSU, BPP): Justin Preece, Justin Elser
- Todorovic Group (OSU, EECS): Sinisa Todorovic, Xu Xu, Ryan Kitchen (Yao Zhou has left the project)
- iPlant-Bisque Group (UCSB): Kris Kvilekval, Dmitri Federov, B.S. Manjunath
Image Annotation Working Group Meetings
Initial Project Meeting: Jan 23, 2015
(Conf. call arranged and hosted on Zoom by Martha Narro, iPlant)
Attendees
- Jaiswal Lab (OSU, BPP): Pankaj Jaiswal, Justin Preece
- Todorovic Group (OSU, EECS): Sinisa Todorovic, Yao Zhou
- iPlant-Bisque Group (UCSB): Kris Kvilekval, Dmitri Federov, B.S. Manjunath
Agenda
- (Re-)Introductions
- Planteome AISO/Bisque proposal review
- Discussion of broad project objectives
- Scheduled next meeting (will be a technical meeting)
Action Items
- Justin send Yao the AISO documentation
- Kris and Dmitry send Yao (and all) pointers to Bisque documentation and Google group.
- Yao: begin to get familiar with Bisque and the AISO Java codebase.
AISO Demo (OSU Only): Tue, Jan 27, 1 PT
Attendees
- Jaiswal Lab (OSU, BPP): Justin Preece
- Todorovic Group (OSU, EECS): Sinisa Todorovic, Yao Zhou, Xu Xu
Agenda
- Demos and technical introductions to AISO and codebase
- Preliminary list of features desired to port to Bisque
Tech Demo Meeting: Thu, Jan 29, noon PT/1p MT
(Conf. call arranged and hosted on Zoom by Martha Narro, iPlant)
Attendees
- Jaiswal Lab (OSU, BPP): Pankaj Jaiswal, Justin Preece
- Todorovic Group (OSU, EECS): Sinisa Todorovic, Yao Zhou, Xu Xu
- iPlant-Bisque Group (UCSB): Kris Kvilekval, Dmitri Federov
Agenda
- Demos and technical introductions to Bisque development environment and AISO functionality
- Second grad student developer (Xu Xu) added from Sinisa's group
- Discuss IGC segmentation algorithm
- Discuss implementation of a Bisque ontology service
- Project planning and timelines
- Discussed possible attendance at May 14-15 Bisque developer workshop at U of Arizona
Tech Dev Meeting: Tue, Feb 10, 1p PT
Attendees
- Jaiswal Lab (OSU, BPP): Justin Preece
- Todorovic Group (OSU, EECS): Yao Zhou, Xu Xu
Agenda
- Discussed approaches to functionality ahead of Bisque tech mtg
- Latency in Bisque's current round-trip approach to Graph Cuts segmentations - is it the round-trip or the graph cuts that takes so long? (JP thinks it's the round trip)
- IGC segmentation algorithm - DCU and Bisque implementations different? (don't know yet)
- Discuss implementation of segmentation module: general use-case
- how to capture and transmit user input (foreground/background markup)
- how to return segmentation results
- emphasized importance of lean data communication, layered segments, overlap capabilities, undo/redo, how to store points sets and annotation data (in generic data structure terms)
- Project planning and timelines: define an iterative dev approach (multiple phases, with modest & clear initial goals)
- will flesh out in mtg w/ Bisque team
Bisque Dev Env Setup Meeting: Thu, Feb 12, 1p MT
(Skype call)
Attendees
- Jaiswal Lab (OSU, BPP): Justin Preece, Justin Elser
- Todorovic Group (OSU, EECS): Yao Zhou, Xu Xu
- iPlant-Bisque Group (UCSB): Kris Kvilekval, Dmitri Federov
Agenda
- Sort out issues in setting up Dev environment for YZ, XX
- Tour of Bisque module XML templates and sample MatLab image annotation script
- Define technical approach and requirements for the first phase of software development.
- Discuss appropriate graph cuts algorithm (will probably use an implementation of Kolmogorov graph cuts
- Discuss latency in Bisque's current round-trip approach to Graph Cuts segmentations
Action Items
- JP - draft requirements documentation for both the initial round of development, as well as more general objectives for the rest of the Planteome Image Annotation efforts
- All - review, discuss, and bicker over said requirements documentation. :)
- YZ, XX, DF, KK - Troubleshoot dev environment setup for YZ and XX.
- Next mtg: TBD
PIA Phase 1 Specification Meeting: Fri, Mar 13, 2p PT
(Planteome Zoom call)
Attendees
- Jaiswal Lab (OSU, BPP): Justin Preece
- Todorovic Group (OSU, EECS): Yao Zhou, Xu Xu
- iPlant-Bisque Group (UCSB): Kris Kvilekval
Agenda
- Revise specs for the first phase of software development
- Continue Dev environment setup with YZ, XX
Action Items
- JP - schedule next meeting for late next week (objectives: put timelines on Phase 1, discuss Phase 2/3 goals)
- KK - complete slimmed-down Bisque dev env deployment package for local installs
- All - think about rough timelines for Phase 1 activities
- YZ, XX, DF, KK - Cont. troubleshooting dev environment setup for YZ and XX.
- Next mtg: TBA (3/20?)
PIA Phase 1 Timeline Meeting: Thu, Apr 23, 10a PT
(Cord 3084)
Attendees
- Jaiswal Lab (OSU, BPP): Justin Preece
- Todorovic Group (OSU, EECS): Sinisa Todorovic, Xu Xu, Ryan Kitchen
Agenda
- Discuss personnel changes: Yao Zhou is leaving the project. Ryan Kitchen (undergrad CS) is joining.
- Discuss timeline for Phase 1
- ST raised the issue of being able to run multiple visual analysis steps in a multi-step "pipeline" within this proposed module. Taken under advisement for future phases; and will factor into the initial architecture as much as possible.
Action Items
- JP - schedule official bi-weekly standing PIA developer meeting
- KK - complete slimmed-down Bisque dev env deployment package for local installs
- Next mtg: TBA
PIA Working Group Bi-Weekly Conf Call: Thu, Apr 30, 2p PT
(Zoom)
Attendees
- Jaiswal Lab (OSU, BPP): Justin Preece
- Todorovic Group (OSU, EECS): Xu Xu, Ryan Kitchen
- UCSB / Bisque Group: Kris Kvilekval
Agenda
- Discuss installation of local Bisque engine package
- Discuss software development timeline and progress
- Prep for UCSB Workshop
Action Items
- XX and JP meet to prep prez and AISO demo for UCSB workshop
PIA Prez Prep Mtg: Tue, May 5, 11:30a PT
(Cord 3084)
Attendees
- Jaiswal Lab (OSU, BPP): Justin Preece
- Todorovic Group (OSU, EECS): Xu Xu, Ryan Kitchen
Agenda
- Prep for UCSB Big Data + Bio-Image Workshop
- Fix AISO installation on MacBook
Action Items
- JP and XX to add and edit assigned slides
PIA Working Group Bi-Weekly Conf Call: Thu, May 28, 3p PT
(Zoom)
Attendees
- Jaiswal Lab (OSU, BPP): Justin Preece, Justin Elser
- Todorovic Group (OSU, EECS): Xu Xu, Ryan Kitchen
- UCSB / Bisque Group: Kris Kvilekval
Agenda
- Software development progress
- Any ongoing issues with the local BisQue engine?
Action Items
- RK is working on the client-side interface
- XX is working on the local engine config and server-side Matlab graph cuts dev
- JP will set up new Planteome repo for PIA source code
- JP will set up new Planteome-Bisque dev mailing list
PIA Working Group Bi-Weekly Conf Call: Thu, Jun 11, 2p PT
(Zoom)
Attendees
- Jaiswal Lab (OSU, BPP): Justin Preece
- Todorovic Group (OSU, EECS): Xu Xu, Ryan Kitchen
- UCSB / Bisque Group: Kris Kvilekval
Agenda
- New mailing list for this group: planteome-bisque-dev@planteome.org
- Software progress
- Start using new Planteome repo: https://github.com/Planteome/planteome-image-annotator
- Commit current work now, please
- Need to get KK, DF added to GitHub team
- Take a look at RK's laptop environment and UI work for our Bisque module
- XX: progress on local engine config and server-side Matlab graph cuts development
- Start using new Planteome repo: https://github.com/Planteome/planteome-image-annotator
- Dennis Stevenson @ NYBG: in situ images
News
- updated Bisque engine coming soon (5.8)...
- good example modules: NuclearDetector3D (latest/greatest), PHFDetector, NuclearFilter (reads Mex for input/pipelining)
- initial module code added to repo; initial matlab scripts coming shortly...
Action Items
- RK adding base module to new repo; adding background blue line markup to existing foreground red line markup
- XX finish engine config, add base Matlab graph cuts scripts to repo
- RK and XX create and confirm round-trip "ping" from client to server within Bisque dev module
PIA Working Group Bi-Weekly Conf Call: Thu, Jun 25, 3p PT
(Zoom)
Attendees
- Jaiswal Lab (OSU, BPP): Justin Preece
- Todorovic Group (OSU, EECS): Xu Xu, Ryan Kitchen
- UCSB / Bisque Group:
Agenda
- Software progress
- Update on Bisque module dev
- adding background blue line markup to existing foreground red line markup
- progress on server-side Matlab graph cuts development
- do we have a successful client-server round-trip?
- Update on Bisque module dev
- Updated Bisque engine (5.8) ETA?
- KK, DF: Accept GitHub team invites, pls
Business
- XX: posted link to Kohli Dynamic Graph Cuts pub (our new algo for graph cuts module)
- RK: stubbed out round-trip from poly-lines to polygon
Action Items
- JP: talk to Nirav about iPlant server space (or explore using CGRB, UCSB BioDev, or EECS space)
- RK: get polygon to return from poly-line input
- XX: Build out Kohli Graph Cuts
PIA Working Group Bi-Weekly Conf Call: Tue, Jul 14, 3p PT
(Zoom)
Attendees
- Jaiswal Lab (OSU, BPP): Justin Preece, Justin Elser
- Todorovic Group (OSU, EECS): Xu Xu, Ryan Kitchen
Agenda
- Software progress
- Update on Bisque module dev
- polygon returning from poly-line input?
- Kohli Graph Cuts progress report
- Updated Bisque engine (5.8) ETA?
- KK, DF: Accept GitHub team invites
- Dev staging space - BioDev seems best
- Update on Bisque module dev
- Dennis: Planteome-Bisque module and your images
Reports
- XX is working on using the existing GraphCutsSegmentation module as a starting codebase, and extending it to include dynamic graph cut functionality. NOTE: This would be a return to using the Kolmogorov algorithm, plus dynamic features. So long as performance is good and scales well, this model should be ok. XX will be conferring with Bisque reps.
- RK having trouble getting local Bisque engine to run Matlab modules; this is needed to test his "round-trip" from client to server and back. He will discuss with KK.
Action Items
- JP: talk to Kris about using UCSB BioDev as our Dev staging server (with build code coming from our GitHub repo)
- RK: troubleshoot local Bisque engine (specifically running Matlab) with KK; there could be an OSU licensing issue
- XX: Discuss dynamic graph cuts system design with DF: pure Matlab, or Matlab + C libraries?
PIA Working Group Bi-Weekly Conf Call: Tue, Aug 18, 3p PT
(Zoom)
Attendees
- Jaiswal Lab (OSU, BPP): Justin Preece, Justin Elser
- Todorovic Group (OSU, EECS): Xu Xu, Ryan Kitchen
- UCSB / Bisque Group: Kris Kvilekval
Agenda
- Dev staging space discussion w/ KK
- KK, DF: Please accept GitHub team invites
- Software progress
- Update on Bisque module dev
- Client-server integration report: RK
- Dynamic Graph Cuts progress report: XX
- Update on Bisque module dev
Reports
- XX has committed the Kohli DynamicGraphCutsSegmentation module to our Github repo, including a compiled algorithm; is currently working to understand interface so as to modify Bisque module to take graph cut output accordingly
- confirmed that our implementation will be a hybrid of Matlab scripts and C++ libraries
- Algorithm author (Kohli) also supplied the starting codebase via public download
- RK is now writing the server-side client interface portion of the module in Python, instead of Matlab
- All agree that this is a good move for flexibility and sustainability
- Discussed with KK: using MCR (Matlab Compiled Runtime) to get around OSU licensing problem; this is needed in order to round-trip test our module interface functionalities.
- KK advised that we will need the OSU Matlab license on our Dev box (just make sure the runtime matches the compiler)
- KK demo'd registering a private Bisque module on their public Bisque engine
- It was decided to set up our environment to do exactly this for integration and performance testing.
In sum, we will create a local Bisque engine (probably on dev.planteome.org) and migrate our centralized dev environment there (pulling from our GitHub repo). Development can still happen on local laptops, but it will also be possible to quickly code and deploy to this Dev engine. Then, we will register and,at intervals, update the version of our module for integration testing in UCSB's environment. Immediate goal: be able to view, run, and test module under development from both a local and a remote Bisque server.
Next step: Focus on a basic integration of graph cuts code and module code (user-facing and Bisque internals).
Action Items
- JP, JE, and RK: set up a local Bisque engine server; migrate module code to it
- RK: register module with UCSB's public server
- XX: continue mods on dynamic graph cuts code
- JP, RK, and XX: Meet to explore interface connections between graph cuts and user-facing module code
PIA Working Group Bi-Weekly Conf Call: Wed, Sep 9, 3p PT
(Zoom)
Attendees
- Jaiswal Lab (OSU, BPP): Justin Preece, Justin Elser
- Todorovic Group (OSU, EECS): Xu Xu, Ryan Kitchen, Sinisa Todorovic
- UCSB / Bisque Group: Kris Kvilekval, Dmitri Federov
Agenda
- Planteome Integrated Dev server update - JP, JE, RK
- KK, DF: Please accept GitHub team invite (this has been hanging out there for a while) - done
- Software progress
- Update on Bisque module dev
- Report on porting client-server piece to Python: RK
- Report on Dynamic Graph Cuts progress (using ImageMatting now - pls explain): XX
- Discuss details of integrating server-side and client-side code: What are the interface points?
Action Items
- Complete deployment and config of Planteome Integrated Dev server - JP, JE, RK
- Define integration points between client and server software - RK, XX
Conferences and Workshops
Please see: Planteome_Outreach