Nov 24th, 2015 Ontology Working Group Meeting: Difference between revisions
(→AmiGO2) |
|||
| (6 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
Date: Tuesday Nov. 24th, 2015 | Date: Tuesday Nov. 24th, 2015 | ||
Time: 9am PST (GMT-8) | Time: 9am PST (GMT-8) | ||
* <sup>Connection details: '''Join from PC, Mac, Linux, iOS or Android: https://zoom.us/j/551476918</sup>''' | * <sup>Connection details: '''Join from PC, Mac, Linux, iOS or Android: https://zoom.us/j/551476918</sup>''' | ||
Attendees: AM, MAL, LC, PJ, JE, CM and EA | * Attendees: AM, MAL, LC, PJ, JE, CM and EA | ||
* Regrets: JD | |||
Regrets: JD | * [[File:Ontology_WG_Meeting_video_11-24-15.mp4|thumb|Ontology WG 11-24-15 video]] | ||
* [[File:Ontology_WG_Meeting_audio_only_11-24-15.m4a|thumb|Ontology WG 10-24-15 audio]] | |||
=Review of recent Planteome Developers' Workshop, Nov 9th-13th, 2015 Corvallis, OR = | =Review of recent Planteome Developers' Workshop, Nov 9th-13th, 2015 Corvallis, OR = | ||
| Line 95: | Line 95: | ||
5. Once term is added, the final term will be posted on GitHub issue and it will be closed. | 5. Once term is added, the final term will be posted on GitHub issue and it will be closed. | ||
=AmiGO2= | =AmiGO2 Demo- TO with CO lentils= | ||
A demonstration version of the AmiGO2.0 Planteome browser has been set up with the TO and the lentil ontology, along with the mappings to the equivalent classes. | * A demonstration version of the AmiGO2.0 Planteome browser has been set up with the TO and the lentil ontology, along with the mappings to the equivalent classes. | ||
* You can view it at: http://dev.planteome.org | |||
* Example: [http://dev.planteome.org/amigo/term/CO_339:0000114 plant height trait] | |||
** Files loaded: dev version of TO: plant-trait-ontology.obo from github and the | |||
* from CM: I made a file called 'ibp-lentil-traits/merged.owl' which is the combination of CO_339 (OWL file?) plus bridging axioms derived from Pankaj's excel spreadsheet. | |||
** [https://raw.githubusercontent.com/Planteome/plant-trait-ontology/master/ext/lentil-traits.owl ibp-lentil-traits/merged.owl] | |||
* The demo is a proof-of-concept for mapping .xls to our browser. Automation will be necessary. Possibly could convert our g-sheet to .csv for the perl script that CM will make to do that automatically. | |||
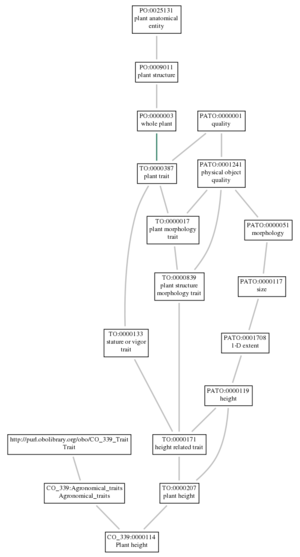
[[File:Visualize.png|thumbnail|center|Plant Height - TO with CO-Lentils]] | |||
=Next Meeting: Tuesday Dec. 8th= | |||
Latest revision as of 00:25, 25 November 2015
Planteome Ontology WG Zoom Meeting:
Date: Tuesday Nov. 24th, 2015 Time: 9am PST (GMT-8)
- Connection details: Join from PC, Mac, Linux, iOS or Android: https://zoom.us/j/551476918
- Attendees: AM, MAL, LC, PJ, JE, CM and EA
- Regrets: JD
- File:Ontology WG Meeting video 11-24-15.mp4
- File:Ontology WG Meeting audio only 11-24-15.m4a
Review of recent Planteome Developers' Workshop, Nov 9th-13th, 2015 Corvallis, OR
- Link to workshop page: http://planteome.org/curation_workshop_2015
- Link to Agenda page: [1]
Attendees:
- Laurel Cooper, Pankaj Jaiswal, Austin Meier, Justin Elser, Justin Preece; Planteome project, OSU
- Barry Smith; OBO Foundry
- Chris Mungall, Seth Carbon; OBO foundry, GO, LBNL
- Marie-Angélique Laporte, Leo Valette, Elizabeth Arnaud (remotely); Bioversity, Crop Ontology Project
- Vilma Hualla, Omar Benites; Bioversity, CIP- Sweet Potato
- Agbona Afolabi- Cassava; Bioversity, IITA
- Karthika Rajendra- Lentil; Bioversity, ICARDA
- Rex Nelson- SoyBase
- Also Sinisa, Eugene, Botong, XuXu
The overall goal is to enrich the Trait Ontology with terms that can be mapped to their equivalents in the various crop specific trait dictionaries.
- Final outcome of the workshop was the mapping of TD terms to the TO, making the TO a true reference ontology, starting with the 5 crops present at the workshop
Shorter term goal (for PAG meeting) is to have a working mappings of the 5 species (lentil, soybean, cassava, sweet potato, banana) that we had at the workshop.
- Need to demonstrate to the breeders how TO can help (need to show the benefits of ontologies.)
- Show them annotation data- needs to be put into the GAF2.0 formatting
- Ideal would be ~5 germplasm phenotypes for each trait in the CO TDs, with the CO mapped to it
- Need germplasm info: Name, source, ID, synonyms, and PHENOTYPES
- note from EA: lentils have not been integrated in the BMS yet
Workflow Details
* Test group- lentil (Karthica), Sweet Potato (Omar & Vilma), cassava (Aflola), banana (Leo), soybean (Rex)
- Work coordinated through GitHub site
- Readme file on each GitHub repo should be filled in with this information
- The Trait Dictionary V.5 xls file - stay intact as a reference
- Create a mapping spreadsheet- similar to “lentil-mapping-TO-CO_339.xls”- leaving the method and scale in, and move to a tab in the google spreadsheet
- The Owl file with proposed mappings from the workshop - MAL will extract mapping from owl files- add to mapping spreadsheet on gsheet. These are the ones the curators selected
- Note: MAL should put the computationally-derived mappings on the gsheet as well. That way we could review them
Mapping Google spreadsheet
- This document is mostly for TO curators, to keep us from altering the TD.xls that is used by the individual crop groups
- MAL extracted Karthica's mappings, and put it in the sheet
- Karthica used parent classes when a term wasn't available. (especially for diseases)
- adding "relationship type" column: default to equivalent, using (s) to indicate subclass
- Lol is grouping them by "class" to better understand the needs within the TO
- Going to compare PJ's mappings to Karthica's mappings
- Karthika used the closest term she could find when she couldn't find the exact term
- PJ promoting the liberal use of synonyms.- Adding synonyms to the TD files eventually would improve the computational mappings?
Curators should Open TO tracker Issues for terms they need added to TO
- Rather not have to create them manually, but it really helps for the discussion of new/complicated terms
- Ideally, TermGenie could create the issue automatically- suggest on tracker-
- Biologist is preferred to OK the terms.
- Complex terms will still require a discussion on GitHub
* Template for Issue tracker for missing terms or ones that need revision:
- Ref to CO term name and ID
- entity and attribute
- proposed TO term name
- Proposed parent in TO
- proposed TO definition
- Optional information:
- link to CO term on CO website
- method and scale (is also on the gsheet)
- literature citation
* EA expresses need for a timeline- so CO contacts for each crop can know what to expect, and when
- The CO curators are busy in the field, but we shouldn't need her assistance until after we work through what we have (Karthica)
- The mapping from their side is 100 terms/week
- The tougher and more time consuming part is fixing the TO.
TermGenie
- TermGenie Instance for TO- Questions and Issues:
- Link to TG instance for TO: TermGenie TO
- TG-TO was not working during the conf call meeting, but now it is, sort of
- Comments, questions etc can be posted on this TO tracker Issue: #361 TermGenie instance for Plant Trait Ontology
- Proposed workflow for requesting terms through TermGenie-TO
1. Sign up for a persona account (for new requesters - from TermGenie page)
2. Add a ticket to the TO tracker, include persona username (email address), requesting TG permission. CM will add you as a user
3. Requests should yield an id automatically, then the term will go into the moderation queue
4. If there are questions, discussion should be on the GitHub issue, not through emails.
5. Once term is added, the final term will be posted on GitHub issue and it will be closed.
AmiGO2 Demo- TO with CO lentils
- A demonstration version of the AmiGO2.0 Planteome browser has been set up with the TO and the lentil ontology, along with the mappings to the equivalent classes.
- You can view it at: http://dev.planteome.org
- Example: plant height trait
- Files loaded: dev version of TO: plant-trait-ontology.obo from github and the
- from CM: I made a file called 'ibp-lentil-traits/merged.owl' which is the combination of CO_339 (OWL file?) plus bridging axioms derived from Pankaj's excel spreadsheet.
- The demo is a proof-of-concept for mapping .xls to our browser. Automation will be necessary. Possibly could convert our g-sheet to .csv for the perl script that CM will make to do that automatically.